Background
Genetic disorders, which are caused by the coding within our DNA, affect millions of people across the world and include conditions such as sickle cell disease and cystic fibrosis.
Natural variations within the DNA base sequence of the human genome, which is approximately 3 billion pairs pairs long, allow us to have different coloured eyes, and hair, for instance. The most common variation in our DNA come from variations in the identity of a DNA base (which can either be G, C, A or T) at a single location in the genome. However, a number of human diseases with a genetic component are caused by similar single base variations and these can either be inherited (e.g. sickle cell disease) or can occur over the course of a person’s lifetime, for example through a cancer-causing mutation. Therefore, being able to identify which base is present at a specific site in the genome is important for research into many genetic diseases, for example in establishing the susceptibility of a person to develop a particular disease or the way that they may respond to a particular drug.
In order to make a chemical sensor to read out the identity of a DNA base, we need to design a molecular probe that produces a response that we can detect when it is exposed to a piece of DNA containing a mutation site. One type of sensor that can be designed is a molecular fluorescence sensor. In this system, the sensing molecule changes the way that it fluoresces when it interacts with a target strand of DNA. To hear more about this sensor development at the University of Birmingham, watch the video below, created by second year Chemistry undergraduate students:
The experiment
AIM
To make a selection of fluorescent solutions, and a molecular fluorescence pH sensor
YOU WILL NEED:

- UV Lamp
- Beakers, conical flasks and glass stirring rods
- Filter paper and funnels
- Spatulas
- Ethanol
- 1M HCl (aq)
- 1M NaOH (aq)
- Spinach leaves
- Non-bio washing powder
- Turmeric (ground)
- Tonic water
- Vitamin B2 (riboflavin) tablets (capsules)
- Distilled water
- Universal indicator pH paper
PROCEDURE
Firstly, make up 5 fluorescent solutions, from materials that can be found in everyday items within the home.
In order to make up an ethanolic chlorophyll solution, crush up ~ 5 g of spinach leaves in 25 mL ethanol. The easiest way to do this is to put the leaves in a sample/sandwich bag with the ethanol, and mash the leaves for several minutes with your hands. Filter this solution, discarding the solid, and collect the green filtrate into a conical flask, labelled ‘ethanolic chlorophyll’.
To make up an ethanolic curcumin solution, dissolve a spatula full (~0.3 g) of turmeric in 25 mL ethanol, and stir the solution well. Filter this solution and collect the yellow filtrate in a labelled conical flask.
To make up an aqueous stilbene solution, dissolve 2 g of non-bio washing power in 50 mL distilled water, stirring carefully so as not to create lots of bubbles. Filter this solution and collect the colourless filtrate into a labelled conical flask.
For the aqueous quinine solution, simply pour 25 mL tonic water into a labelled conical flask.
Finally, in order to make up an aqueous riboflavin solution, empty the contents of 1 vitamin B2 capsule into a beaker containing 50 mL distilled water, and stir the mixture thoroughly (note – much of the content will not dissolve well). Filter the mixture and collect the yellow filtrate into a labelled conical flask.
Pipette a few mLs of each solution into test tubes/small vials and look at them under UV light. Look at a sample of water and ethanol also, as a control. Record the colour of the solutions in daylight and also when they fluorescence under UV light.
Now you can make a solution that can be used as a fluorescence sensor; to do this we need to use a molecule that responds to an input, and change the way in which it fluoresces.
Take 2 test tubes and add 5 mL of distilled water and 1mL of the aqueous riboflavin solution to each, and mix them with a glass stirring rod. Test the pH of the solutions with universal indicator paper. With one of the test tubes, add a few drops of 1 M HCl (aq) and stir, until the pH of the solution reaches 1. Now look at the two solutions under a UV lamp, and compare the fluorescence. You can reverse the change by adding 1 M NaOH (aq) dropwise, bringing the pH back to 7. You can also see what happens to the fluorescence when the pH reaches 14.
Questions
- Looking at the structures of the molecules that are fluorescent, what do you notice that they all have in common?

- What are the fluorescent molecules doing when you shine UV light on them? What type (wavelengths) of light are they absorbing? What type of light are they emitting?
- Why do you think fluorescent stilbene molecules are added as optical brighteners to washing powders? Paper also contains similar optical brighteners, have a look at some white paper in the dark under the UV lamp.
- Is fluorescence a fast process? Does the fluorescence stop when you switch off the UV lamp?
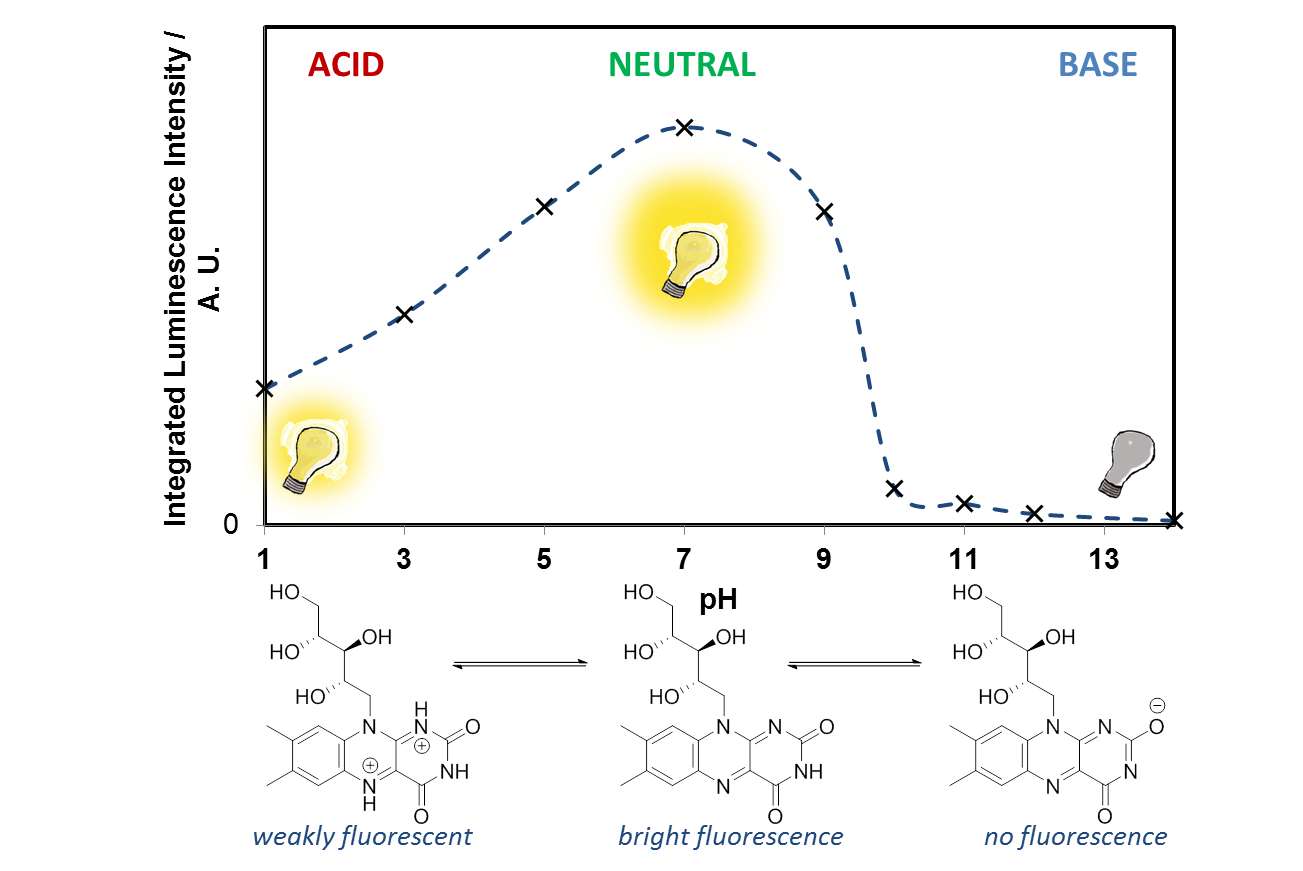
- Look at the two riboflavin solutions, what effect does the pH have on the fluorescence? Is the effect reversible? Looking at the molecular structure of riboflavin, can you suggest what is happening at low and high pH? (hint – you are affecting the N atoms in the ring system) Why does this affect the fluorescence?
To find out more about acids, bases, and pH, take a look at the University of Birmingham resource found here.
Going Further
It is easy to see with your eyes that the fluorescence is different at pH 7, compared with the riboflavin solution at pH 14, or pH 1, but it is not so easy to see the difference between these two solutions. It is also possible to analyse this more accurately using a technique called photoluminescence spectroscopy.
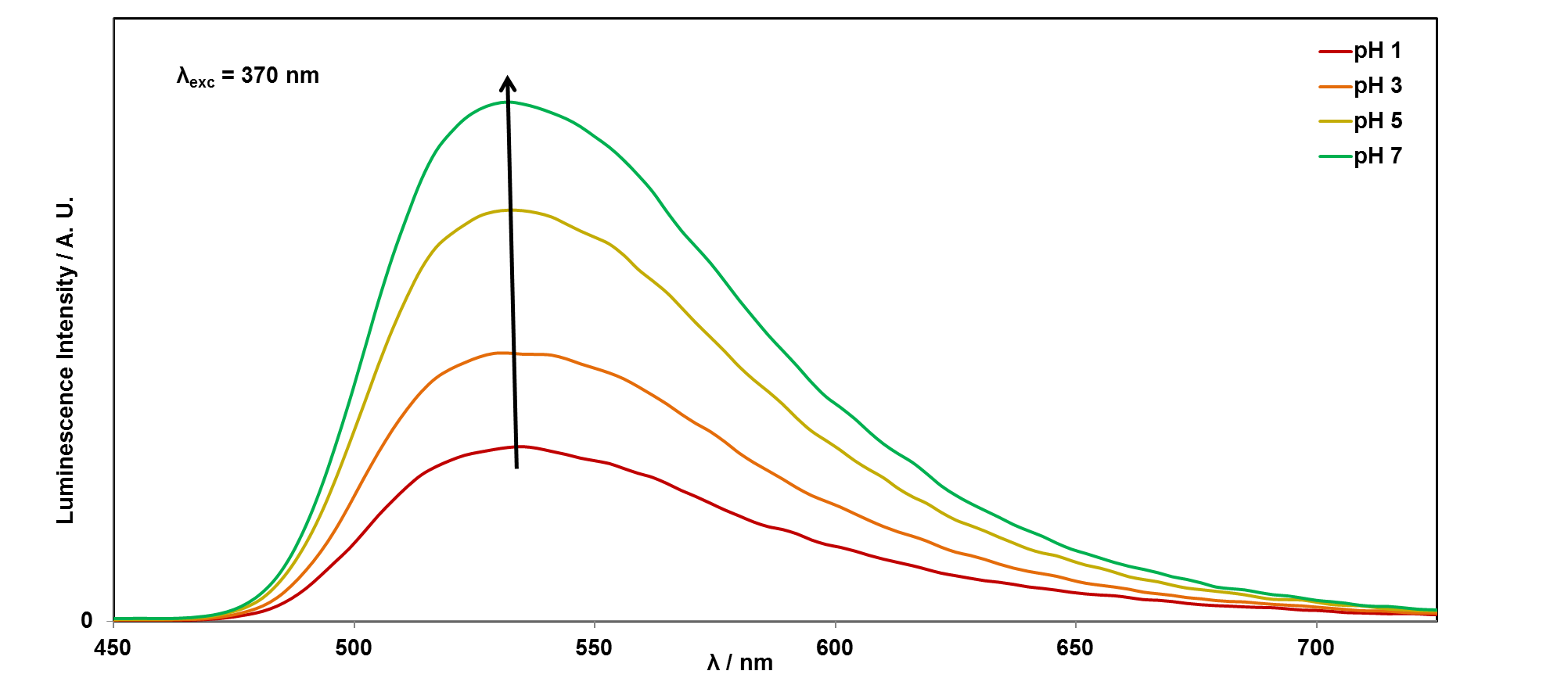
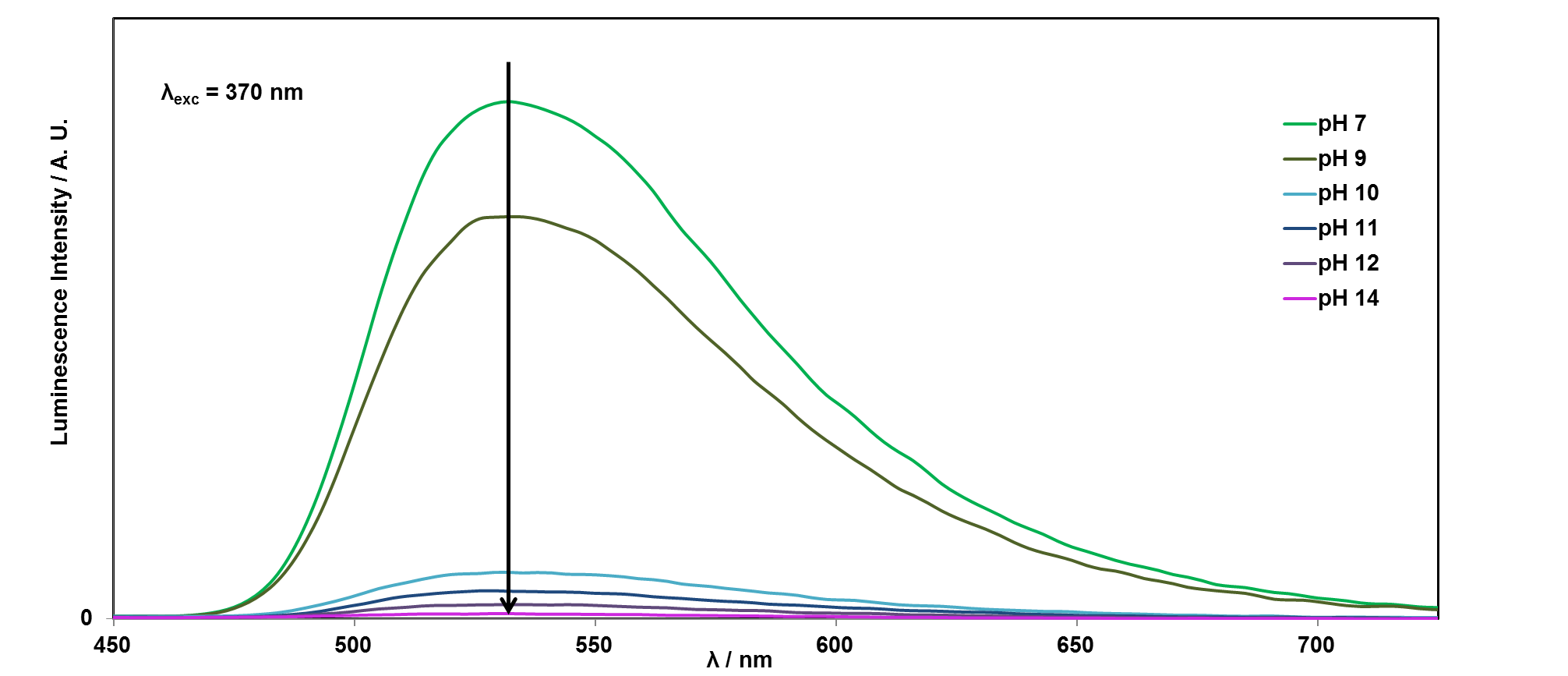
If you would like to plot these graphs yourself, you can download the data here. We can also plot the emission intensity against the pH of the solution, to see how the sensor responds over the pH range 1-14:
In the research lab
In the above experiment, fluorescent molecular sensors are used to detect changes in pH. In the School of Chemistry at the University of Birmingham, researchers working with Jim Tucker have been using a similar concept to develop sensors that detect base changes (SNPs) in DNA. To be able to detect which SNP is present in a section of DNA, the researchers first make a short strand of DNA that will match with one form of the SNP, that has a fluorescent molecule in place of one of the bases; this is the fluorescent sensor (probe):
When this fluorescence sensor is added to the DNA, it binds with the DNA to form a double helix. The fluorescence of the sensor will be different depending upon whether the DNA bases matched or mis-matched, because the local environment around the fluorescent molecule changes. Therefore, the fluorescence output changes with the SNP input, and can be used to detect various diseases.
To find out more about this research click here.
Publications about fluorescence sensors from the University of Birmingham
Abstract above reprinted with permission, Copyright (2015) American Chemical Society. http://pubs.acs.org/doi/pdf/10.1021/acschembio.5b00796
Abstract above reprinted under CC-BY license. To access this paper click here.

This work is licensed under a Creative Commons Attribution 4.0 International License.


