Background
DNA (deoxyribonucleic acid) is present in the living cells of animals and plants, and contains the genetic code that determines the make-up of every single protein of an organism. DNA is coiled up in our chromasomes, and the complete set of genetic material is called our genome.

Consider your genome as a book; it is 3 billion base-pairs long, with a start, a middle, and an end, and contains all the information to code every cell in our body how to work. Geneticists want to be able to ‘read’ this code. All current DNA sequencing techniques used to read this code have to break the genome into millions of tiny fragments, and then try to put them all back together using supercomputers. This is a bit like cutting up a book into its individual words, and then trying to piece the book back together with no prior knowledge of the story being told. Some words will only occur once in the book, but many will also repeat several times throughout the story. In order to tell the story, you need context; a long-range map of the story that tells you how many times the repeating text occurs, and the distance between repeats. Researchers at the University of Birmingham working with Robert Neely make genome maps, i.e. DNA barcodes, in order to read out information on the nature of the landscape of the genome.

To see how this is useful, lets look further at the book analagy: taking two books, Alice’s Adventures in Wonderland (Lewis Carroll) and Emma (Jane Austen), if we cut each book up into all its individual words it would be difficult to tell the two apart; both contain millions of words. However, if we look at just one word, the word ‘think‘, this appears 53 times within Alice’s Adventures in Wonderland, and 384 times in Emma. If we were to line up all the letters of all the words within each book, and then just mark the word ‘think’ with a black line, the two ‘barcodes’ created would be easy to tell apart.
Experiment
DNA is a polymer made by the condensation polymerisation of repeating units (monomers) called nucleotides. The DNA of strawberries and bananas can be extracted relatively easily; firstly the fruit cells need breaking apart from one another, and secondly the cell membrane and the membrane around the nucleus need to be broken up, in order to release the DNA. The membranes are made up of fatty molecules, and can be disrupted using simple detergents, such as washing-up liquid.
AIM
To extract the DNA from the cells of strawberries and bananas
YOU WILL NEED

- Bananas/ strawberries
- Sodium chloride (or table salt)
- Washing-up liquid
- Distilled water
- Ethanol (cooled over ice, or in the fridge or freezer)
- Sample bags/sealable sandwich bags
- Fruit knife
- Small sieve
- Glass beakers and stirring rods
- Plastic Pasteur pipettes
- Tweezers/wooden skewers
PROCEDURE
To make the extraction mixture: Weigh out 3 g of NaCl (s) and add to a 150 mL glass beaker. Add 10 mL of washing-up liquid (2 tsp) and make up to 100 mL with distilled water. Gently stir the solution to mix it together, without forming too many bubbles.
To extract the DNA: Take 1 strawberry and/or a similar sized piece of banana (peeled), cut it up into small pieces and place into a plastic bag, with 50 mL of the extraction mixture. Seal the bag with minimal air in it, mash the fruit inside the bag by squeezing it (without bursting the bag!) for 5-10 min, to form a relatively smooth puree. Filter the puree through a sieve lined with kitchen towel.
Take 20 mL ice-cold ethanol in a beaker, and carefully drop 2 mL of the fruit mixture into this; do not shake it. After a few minutes you should be able to see some insoluble material precipitating out in the ethanol, which you can remove from the solution with tweezers.

QUESTIONS
- Why is cold ethanol used to precipitate out the DNA?
- What does the DNA look like?
- What effect does the detergent have on the way the puree looks? – try to mash the strawberries in 50 mL water / 1.5 g salt. Does it look different?
- When salt is added, the ions help to create an environment where the DNA clumps together, and is easier to see and extract from the ethanol – why do you think this is?
Going further
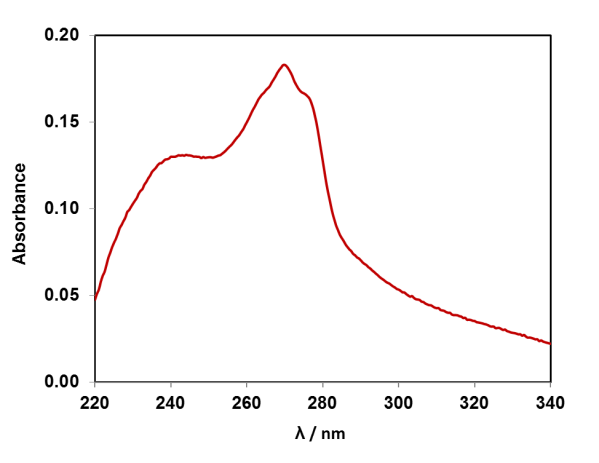
If we dissolve the DNA in water, we can take a UV-Vis absorption spectrum of the solution, as the DNA absorbs light in the UV region of the electromagnetic spectrum. The four DNA bases (A, T, G, C) contain aromatic rings with alternating double-single C-C bonds, like benzene. The pi-electrons in these systems absorb UV light. Each of the four DNA bases has a slightly different absorption spectrum, and the spectrum of DNA is the average of them. Compare the UV-Vis absorption spectrum of strawberry DNA above to the four DNA bases, can you spot the adenine feature? (Data available for plotting in resource pack).
In the research lab
In the School of Chemistry at the University of Birmingham, researchers extract DNA in a similar fashion, in order to label it with markers to create a genome map, or a ‘DNA barcode’. The DNA is extracted from cells using a detergent-type molecule to disrupt the cell membrane, and precipitation of the DNA in ethanol separates it from the rest of the cell matter. (Usually this DNA is further purified to remove any proteins that may still be present.)
The DNA is labelled using enzymes (proteins that catalyse very specific biochemical reactions) as molecular machines, to modify the chemistry of extracted DNA, in order to tag sequences of bases with fluorescent markers.

In bacteria, certain enzymes modify DNA to protect itself against the invasion of foreign DNA from viruses; to find out more about this click here. These DNA-modifying enzymes are used to label specific sites in the DNA. The enzyme binds to a specific base sequence, i.e. a specific piece of code within the DNA molecule, everytime it appears. When a cofactor (a non-protein molecule that reacts with the enzyme and the DNA to complete the DNA modification reaction) is added, this also binds to the enzyme. The enzyme ‘locks’ the correct bit of DNA code in place with the cofactor, so that a nucleophilic substitution reaction can occur, joining the DNA with a covalent bond to a fluorescent tag.
We can use this technique to create ‘DNA barcodes’ that we can see under a fluorescent microscope.

Once the DNA has been labelled, the long strands need to be untangled and straightened out. If we just deposit the DNA onto glass slides from droplets of solution, they remain all tangled up like spaghetti. In order to do this, researches at the University of Birmingham have developed a method to ‘comb’ the DNA into straight strands, and then look at them under a fluorescence microscope to see the markers, like a barcode. Droplets of water that have picograms of DNA dissolved in them (1 pg = 10-12 g, which means there are a million million pg in 1 g!) are dropped onto a glass slide from a pipette. Special glass slides are used that are coated in a polymer. The DNA tends to unwrap slightly at one end and tether to the polymer coating. Then as the droplet is dragged across the slide, the DNA is stretched out, and finally tethers on the other end, in a ‘combing’ process.
These DNA barcodes can then be screened very quickly, and used as a new fast method for detecting and identifying disease-causing organisms, such as viruses and bacteria.
Find out more



Edited by Emily Hanover 05/07/2022
This work is licensed under a Creative Commons Attribution 4.0 International License.